Difference between revisions of "User talk:Tindle Lisa/My sandbox Support page v2"
From Bioblast
Tindle Lisa (talk | contribs) |
Tindle Lisa (talk | contribs) |
||
| Line 1: | Line 1: | ||
{{Template:OROBOROS support page name}} | |||
{{NextGen-O2k H2020-support}} | |||
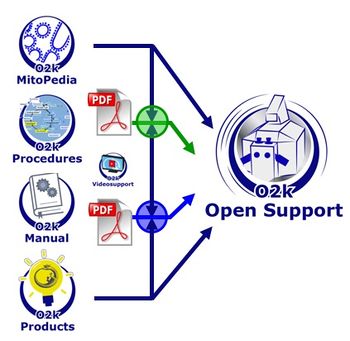
[[File:O2k-support system.jpg|right|350px|Oroboros support system]] | |||
:::::::::::: <big>''Currently Asked Questions - »'''[[O2k-Open Support alert]]'''« </big> | |||
__TOC__ | |||
== O2k-Quality Control == | |||
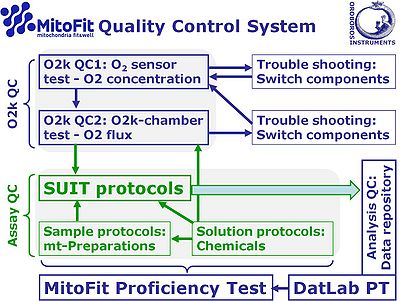
[[File:MitoFit-QCS.jpg|right|400px|MitoFit-QCS]] | |||
:::: Many commonly raised issues of O2k-technical support are solved in the newest version of [[DatLab]]. | |||
:::: The '''O<sub>2</sub> sensor and O2k chamber tests''' are important to eliminate instrumental artefacts (instrumental quality control, QC), distinguished from assay QC addressing problems of [[solution protocols]] (preparation of chemicals), [[sample]] preparation protocols (such as [[mitochondrial preparations]]), and [[SUIT protocols]]. | |||
::::# Clean O2k-chambers and contamination-free incubation medium are required. | |||
::::# Analysis of DatLab files recorded in instrumental QC tests is essential. | |||
::::# Biological experiments are not suitable for trouble shooting. | |||
=== 1. O<sub>2</sub> sensor test === | |||
:::: The [[OroboPOS|OroboPOS]] (polarographic oxygen sensor) requires regular service at intervals - these may be more than 1 year. SOPs are available to determine if a sensor service is required at an earlier or later date: | |||
::::» O<sub>2</sub> sensor test: [[Oxygen_sensor_test |'''QC1: Oxygen sensor test''']] | |||
=== 2. O2k-chamber test === | |||
:::: If all quality control criteria of the O<sub>2</sub> sensor test are met, the operator can be assured that the quality of the sensor signal is acceptable. Next, the quality of the O2k-chamber assembly has to be tested, described in detail: | |||
::::» O2k-chamber test: [[Oxygen flux - instrumental background |'''QC2: Instrumental O<sub>2</sub> background test''']] | |||
== O2k-Open Support: Installation and startup support session == | |||
:::: Each O2k-Package includes a remote installation and startup support session with an Oroboros expert. This online support session will focus on: | |||
::::* Instrumental setup | |||
::::* Quality control | |||
::::* Discussion with our expert | |||
:::: Installation and startup support can be scheduled during any time within the O2k-Warranty period (max. 2 years) | |||
:::::» '''[https://www.oroboros.at/index.php/support-session-request-form/ Register for your session]''' | |||
:::: Upon special request and subject to an additional fee, Oroboros Instruments offers a start-up visit by a scientific application specialist. | |||
:::: | :::::» '''[[O2k-Start up and Training]]''' | ||
: | [[File:O2k-Support.jpg|right|200px|O2k-Open Support]] | ||
=== | == O2k-Open Support agreement == | ||
:::: | :::: The Oroboros experts will be happy to help in using our O2k-Open Support system. | ||
::::» [[Search for defective O2k components]] | |||
:::: '''»''' In case you need further technical support and for your valuable feedback, please contact [https://www.oroboros.at/index.php/o2k-technical-support '''O2k-Tech Support'''] | |||
::::» [[O2k-Open Support agreement]] | |||
::::» [[O2k repair]] | |||
[[File:O2k-Network.png|left|100px|O2k-Network|link=O2k-Network]] | |||
== The O2k-Network == | |||
:::: | :::: The Oroboros O2k-Network serves to connect and support: Contact for advice on applications of the O2k and high-resolution respirometry - the global network in mitochondrial physiology. | ||
::::: '''»''' The O2k-Network Labs: [[O2k-Network|» O2k-Network]] | |||
[[ | |||
::::: '''»''' [[O2k-Network_discussion_forum|O2k-Network discussion forum]] | |||
:::: | |||
:::: '''»''' [[Oroboros Laboratories|Oroboros O2k-Laboratory]] - open for innovation and cooperation. | |||
:::: Oroboros | :::: '''»''' [[Oroboros_Science_Scholarship|Oroboros Science Scholarships]] - are offered for focused research projects on mitochondrial physiology and high-resolution respirometry | ||
[[ | |||
[[Category:O2k-Open Support]] | |||
Revision as of 14:36, 20 May 2021
 |
User talk:Tindle Lisa/My sandbox Support page v2 |
- Currently Asked Questions - »O2k-Open Support alert«
O2k-Quality Control
- Many commonly raised issues of O2k-technical support are solved in the newest version of DatLab.
- The O2 sensor and O2k chamber tests are important to eliminate instrumental artefacts (instrumental quality control, QC), distinguished from assay QC addressing problems of solution protocols (preparation of chemicals), sample preparation protocols (such as mitochondrial preparations), and SUIT protocols.
- Clean O2k-chambers and contamination-free incubation medium are required.
- Analysis of DatLab files recorded in instrumental QC tests is essential.
- Biological experiments are not suitable for trouble shooting.
- The O2 sensor and O2k chamber tests are important to eliminate instrumental artefacts (instrumental quality control, QC), distinguished from assay QC addressing problems of solution protocols (preparation of chemicals), sample preparation protocols (such as mitochondrial preparations), and SUIT protocols.
1. O2 sensor test
- The OroboPOS (polarographic oxygen sensor) requires regular service at intervals - these may be more than 1 year. SOPs are available to determine if a sensor service is required at an earlier or later date:
- » O2 sensor test: QC1: Oxygen sensor test
2. O2k-chamber test
- If all quality control criteria of the O2 sensor test are met, the operator can be assured that the quality of the sensor signal is acceptable. Next, the quality of the O2k-chamber assembly has to be tested, described in detail:
- » O2k-chamber test: QC2: Instrumental O2 background test
O2k-Open Support: Installation and startup support session
- Each O2k-Package includes a remote installation and startup support session with an Oroboros expert. This online support session will focus on:
- Instrumental setup
- Quality control
- Discussion with our expert
- Each O2k-Package includes a remote installation and startup support session with an Oroboros expert. This online support session will focus on:
- Installation and startup support can be scheduled during any time within the O2k-Warranty period (max. 2 years)
- Upon special request and subject to an additional fee, Oroboros Instruments offers a start-up visit by a scientific application specialist.
O2k-Open Support agreement
- The Oroboros experts will be happy to help in using our O2k-Open Support system.
- » Search for defective O2k components
- » In case you need further technical support and for your valuable feedback, please contact O2k-Tech Support
- » O2k-Open Support agreement
- » O2k repair
The O2k-Network
- The Oroboros O2k-Network serves to connect and support: Contact for advice on applications of the O2k and high-resolution respirometry - the global network in mitochondrial physiology.
- » The O2k-Network Labs: » O2k-Network
- » Oroboros O2k-Laboratory - open for innovation and cooperation.
- » Oroboros Science Scholarships - are offered for focused research projects on mitochondrial physiology and high-resolution respirometry