Difference between revisions of "PGM-pathway control state"
| Line 35: | Line 35: | ||
::::* DL-Protocol for permeabilized fibers (pfi):[[SUIT-001 O2 pfi D002]] | ::::* DL-Protocol for permeabilized fibers (pfi):[[SUIT-001 O2 pfi D002]] | ||
::::* DL-Protocol for permeabilized cells (pce): [[SUIT-001 O2 ce-pce D003]] | ::::* DL-Protocol for permeabilized cells (pce): [[SUIT-001 O2 ce-pce D003]] | ||
::::* DL-Protocol for permeabilized | ::::* DL-Protocol for permeabilized PBMC and PLT: [[SUIT-001 O2 ce-pce D004]] | ||
:::*[[SUIT-012]] | :::*[[SUIT-012]] | ||
== Discussion == | == Discussion == | ||
Revision as of 09:48, 3 March 2020
Description
PGM: Pyruvate & Glutamate & Malate.
MitoPathway control state: NADH Electron transfer-pathway state
SUIT protocol: SUIT-001, SUIT-012, SUIT-014
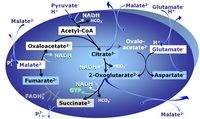
Pyruvate (P) is oxidatively decarboxylated to acetyl-CoA and CO2, yielding NADH catalyzed by pyruvate dehydrogenase. Malate (M) is oxidized to oxaloacetate by mt-malate dehydrogenase located in the mitochondrial matrix. Condensation of oxaloacate with acetyl-CoA yields citrate (citrate synthase). Glutamate&malate is a substrate combination supporting an N-linked pathway control state, when glutamate is transported into the mt-matrix via the glutamate-aspartate carrier and reacts with oxaloacetate in the transaminase reaction to form aspartate and oxoglutarate. Glutamate as the sole substrate is transported by the electroneutral glutamate-/OH- exchanger, and is oxidized in the mitochondrial matrix by glutamate dehydrogenase to α-ketoglutarate ( 2-oxoglutarate), representing the glutamate anaplerotic pathway control state. 2-oxoglutarate (α-ketoglutarate) is formed from isocitrate (isocitrate dehydrogenase, from oxaloacetate and glutamate by the transaminase, and from glutamate by the glutamate dehydrogenase.
Abbreviation: PGM
Reference: Gnaiger 2014 MitoPathways - Chapter 5.3
MitoPedia concepts:
SUIT state
PGML
PGM pathway in the LEAK state can be evaluated in the following SUIT protocols:
PGMP
PGM pathway in the OXPHOS state can be evaluated in the following SUIT protocols:
-
- DL-Protocol for permeabilized fibers (pfi): SUIT-008 O2 pfi D014
- DL-Protocol for permeabilized cells (pce): SUIT-008 O2 ce-pce D025
-
PGME
PGM pathway in the ET state can be evaluated in the following SUIT protocols:
-
- DL-Protocol for isolated mitochondria and tissue homogenate (mt):SUIT-001 O2 mt D001
- DL-Protocol for permeabilized fibers (pfi):SUIT-001 O2 pfi D002
- DL-Protocol for permeabilized cells (pce): SUIT-001 O2 ce-pce D003
- DL-Protocol for permeabilized PBMC and PLT: SUIT-001 O2 ce-pce D004
-